Tip
All input files can be downloaded: Files.
md
This keyword controls the molecular dynamics (MD) simulation details.
Options
- type
Value
NVEMicrocanonical ensemble, constant number of particles, volume, and energyNVTCanonical ensemble, constant number of particles, volume, and temperaturefep-NVEFree energy perturbation calculation for microcanonical ensemblefep-NVTFree energy perturbation calculation for canonical ensembleDefault
NVEDefine the MD simulation type.
- dt
Value
A real number
Default
0.0005The time step in ps. For systems not containing hydrogen atoms,
dtcan be set to0.001ps.
- num_steps
Value
An integer
Default
10The number of MD steps to be run.
The total simulation time is
dt``*\ ``num_stepsps.
- output_freq
Value
An integer
Default
1The number of steps that MD information is output. The trajectory, velocity, and gradient will be output to disk.
- temp
Value
A real number
Default
273.15Define the initial temperature for NVE simulation, and the targeted temperature for NVT simulation.
- temp_nhc_length
Value
An integer
Default
5The length of Nose-Hoover chain to control the temperature. Usually an integer larger than 2 is enough.
- temp_nhc_tau
Value
A real number
Default
0.5The time-constant of Nose-Hoover chain to control the temperature in ps.
- vel_fn
Value
A file name
Default
None
If this is given, the initial velocities will be set from this XYZ file (in Angstrom/ps). Otherwise, velocities will be generated from Boltzmann distribution. For example:
1md 2 vel_fn mol-vel.xyz 3end
Initial velocities will be read from
mol-vel.xyz.
- max_dr
Value
A real number
Default
5The maximum allowed atomic displacement per step in Angstrom. If an atom mover more than
max_drin a step, Qbics will stop the MD simulation since the system may crash due to the too fast motion.
- plumed_fn
Value
A file name
Default
None
If this is given, Qbics will apply enhanced sampling algorithms of PLUMED defined in this file. For PLUMED input file grammar, please refer to its PLUMED manual.
Theoretical Background
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations are a computational method to study the time evolution of a system of interacting particles. In MD simulations, the forces between particles are calculated using a potential energy function, and the equations of motion are solved to obtain the trajectory of the particles.
In Qbics, the equations of motions are solved using the velocity-Verlet algorithm, which is a symplectic integrator, meaning that it conserves the total energy of the system over long time. The forces can be evaluated on several levels of theory, including DFT, MM, NDDO, xTB, DFT/MM, NDDO/MM, and xTB/MM.
During MD simulations, Qbics will output several files:
x-md-traj.xyz: The trajectory of the system in XYZ format.x-md-vel.xyz: The velocity of the system in XYZ format.x-md-grad.xyz: The gradient of the system in XYZ format. Note that force is the negative of gradient.x-md-log.txt: The log file of the MD simulation, containing the step, time, energy components, temperature, and other information.x-md-pressure.txt: The pressures of the MD simulation, containing the step, time, cell volume, and pressure tensor. If the system is not periodic, this file will not be generated.
If plumed_fn is given, Qbics will also output some data defined in this file.
Before doing MD, it is strongly recommended to perform a geometry optimization to obtain a stable initial configuration.
Each ensemble has its own conserved quantity.
For NVE, the total energy is conserved:
Here \(E\) is the total energy, \(K\) is the kinetic energy, and \(V\) is the potential energy.
For example, if you do an NVE MD simulation, you can check the total energy in file x-md-log.txt:
1 # Time Total Kinetic Potential Temp Cost
2 ps Hartree Hartree Hartree Kelvin second
3 0 0.00000 -34.44404963 0.02280104 -34.46685067 300.00000000 0.257
4 20 0.01000 -34.44402464 0.01224859 -34.45627323 161.15837759 0.260
5 40 0.02000 -34.44400898 0.01020550 -34.45421449 134.27682685 0.222
6 60 0.03000 -34.44401439 0.01628312 -34.46029751 214.24190576 0.220
7 80 0.04000 -34.44402295 0.02160569 -34.46562864 284.27246073 0.279
8 100 0.05000 -34.44403491 0.03125266 -34.47528758 411.20057981 0.248
9 120 0.06000 -34.44402767 0.02061378 -34.46464145 271.22164167 0.228
10 140 0.07000 -34.44403668 0.02599444 -34.47003112 342.01656058 0.226
11 160 0.08000 -34.44402275 0.01897475 -34.46299751 249.65643218 0.224
For NVT, the total “virtual” energy is conserved:
Here \(n_\text{NHC}\) is the Nose-Hoover chain length nhc_temp_length, \(k\) is the Boltzmann constant, \(T\) is the temperature, \(\eta_j\) and \(p_{\eta_j}\) are the thermostat variables (“position” and “momentum”), and \(Q_j\) is the thermostat mass (determined automatically by Qbics).
For example, if you do an NVT MD simulation, you can check the ConstQ in file x-md-log.txt:
1 # Time Total Kinetic Potential Temp ConstQ Cost
2 ps Hartree Hartree Hartree Kelvin Hartree second
3 0 0.00000 -34.44404963 0.02280104 -34.46685067 300.00000000 -34.44404963 0.000
4 20 0.01000 -34.44404123 0.01734433 -34.46138557 228.20455596 -34.44404256 0.293
5 40 0.02000 -34.44401539 0.01535660 -34.45937199 202.05137626 -34.44402161 0.230
6 60 0.03000 -34.44399789 0.01914301 -34.46314090 251.87027197 -34.44401529 0.213
7 80 0.04000 -34.44401533 0.03373382 -34.47774915 443.84591789 -34.44404524 0.303
8 100 0.05000 -34.44399074 0.01808475 -34.46207549 237.94640025 -34.44402711 0.222
9 120 0.06000 -34.44400165 0.03088255 -34.47488420 406.33083745 -34.44404511 0.218
10 140 0.07000 -34.44397432 0.01350937 -34.45748370 177.74683071 -34.44402030 0.327
11 160 0.08000 -34.44397272 0.01886731 -34.46284002 248.24278233 -34.44402353 0.222
12 180 0.09000 -34.44398418 0.03438731 -34.47837149 452.44411562 -34.44404296 0.225
In all cases, if the conserved quantity is not conserved, it means that the simulation is unstable. You may need:
Reduce the time step;
Do a geometry optimization to obtain a stable initial configuration;
Check if the potential energy function (quantum chemistry or force field parameters) is reasonable for your system.
However, if plumed_fn is given, the conserved quantity may NOT be conserved, since the enhanced sampling algorithm may change the total energy.
Nose-Hoover Chain algorithm
The Nose-Hoover chain (NHC) algorithm is a method to control the temperature in MD simulations. It is a generalization of the Nose-Hoover thermostat. The NHC is a series of thermostats that are connected to each other. The first thermostat controls the temperature of the system, and the subsequent thermostats control the temperature of the previous thermostat.
The length of NHC is controlled by temp_nhc_length. It should be larger than 2. If temp_nhc_length is set to 1, the system may not have a correct canonical distribution of velocities.
Input Examples
Example: Dynamic Effects for Carbocation Reaction
Consider the following reaction:
We want to know which group is more likely to migrate. Thus, we will do a 200 fs NVT MD simulation from the optimized structure of this carbocation to see what will happen. We will repeat the MD for 20 times using a shell script run.sh. Here we use xTB for its speed. You can also use DFT if you have enough time. The input files are shown below:
1mol
2 C -0.90849 0.15356 -0.48118
3 C -0.18840 -0.79035 0.24223
4 C -0.79661 -1.92884 0.73482
5 C -2.15434 -2.10431 0.51590
6 C -2.88201 -1.15101 -0.17642
7 C -2.26735 -0.01407 -0.67971
8 H -0.22972 -2.66722 1.27887
9 H -2.64692 -2.98493 0.89733
10 H -3.93966 -1.29437 -0.33076
11 H -2.84788 0.72259 -1.21038
12 C 1.26411 -0.39651 0.38023
13 C -0.02903 1.30229 -0.93562
14 C 1.67317 -0.22550 1.85240
15 H 1.83624 -1.21169 2.27913
16 H 2.60525 0.32605 1.95669
17 H 0.88426 0.23363 2.45602
18 C -0.71228 2.64784 -0.65267
19 H -0.02047 3.48874 -0.70508
20 H -1.46470 2.82054 -1.41711
21 H -1.26222 2.63862 0.29289
22 C 1.31204 0.95650 -0.38570
23 H 2.18545 1.61818 -0.54700
24 C 2.13967 -1.43173 -0.30477
25 H 1.61435 -2.09539 -0.97595
26 C 3.43556 -1.57292 -0.10516
27 H 3.99105 -2.34108 -0.61298
28 H 4.00084 -0.94936 0.56664
29 C 0.38001 1.16971 -2.43593
30 C 1.13470 2.39986 -2.94354
31 C -0.80675 0.90408 -3.35872
32 H 1.04783 0.30446 -2.50076
33 H 1.96481 2.68234 -2.30028
34 H 1.54266 2.18219 -3.92659
35 H 0.47031 3.25225 -3.05019
36 H -1.34333 0.00481 -3.07250
37 H -1.49736 1.74331 -3.36799
38 H -0.44263 0.76667 -4.37283
39end
40
41basis
42 def2-svp
43end
44
45pseudopotential
46 def2-ecp
47end
48
49scf
50 charge +1
51 spin2p1 1
52end
53
54xtb
55 chrg +1
56end
57
58md
59 type nvt
60 dt 0.001 # 0.001 ps = 1 fs
61 num_steps 200 # 0.001*200 = 0.2 ps. In real simultion, you may need > 1 ps.
62 temp 300
63 temp_nhc_length 5
64 temp_nhc_tau 0.5
65 output_freq 10 # 0.001*10 = 10 fs
66end
67
68task
69 md xtb
70 # md m06 # If you want to use DFT.
71end
1for i in {1..20}; do
2 fn=md-1-$i
3 cp md-1.inp ${fn}.inp
4 qbics ${fn}.inp -n 8 > ${fn}.out
5done
By bash run.sh, you can run 20 MD simulations. The output files will be md-1-1-md-traj.xyz, etc. We observe that both the isopropyl and vinyl group can migrate. For example:
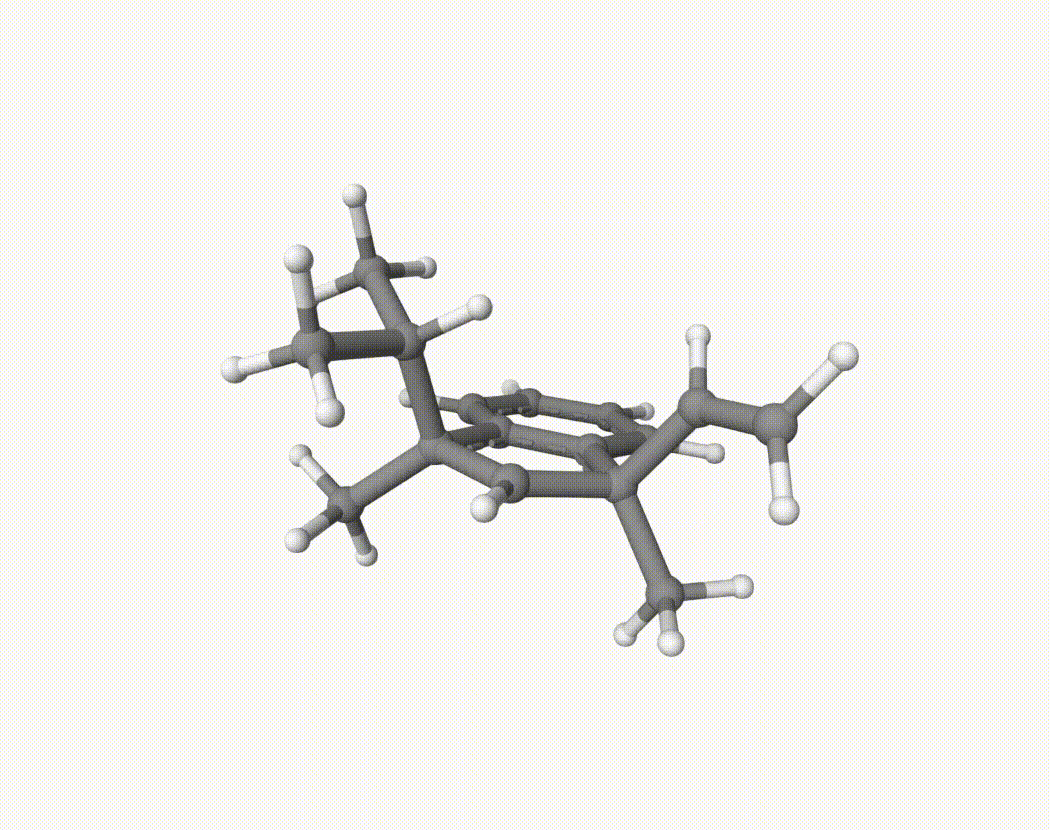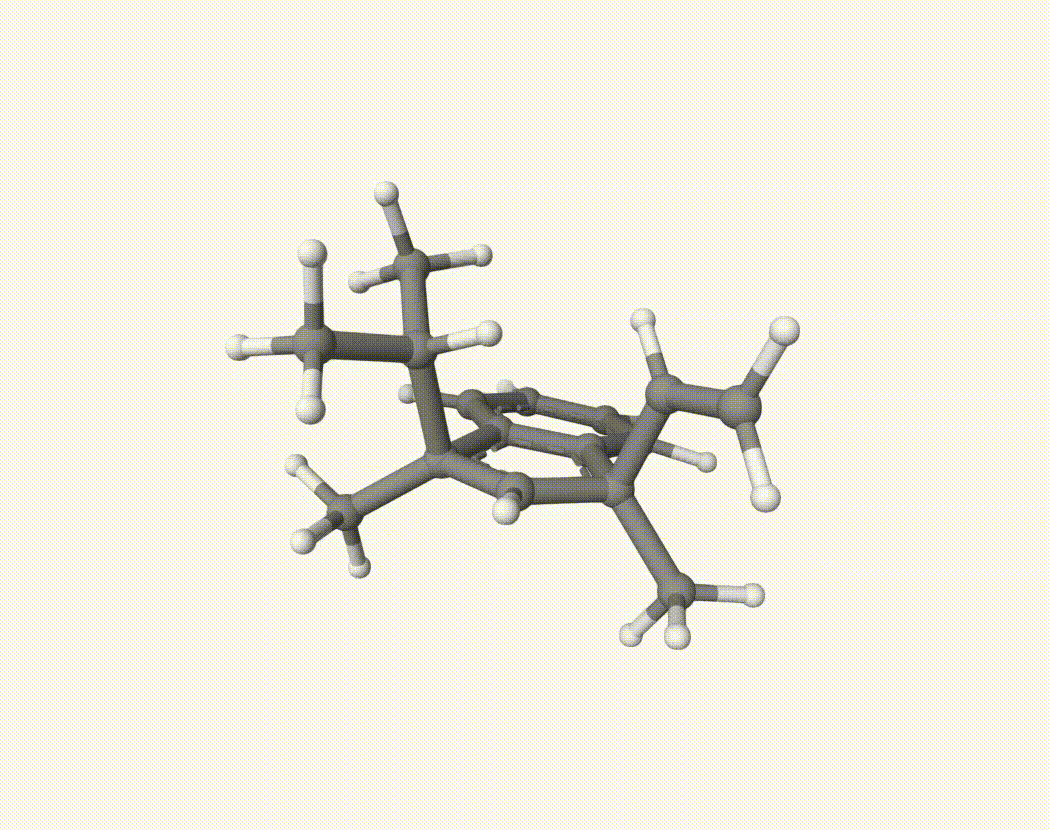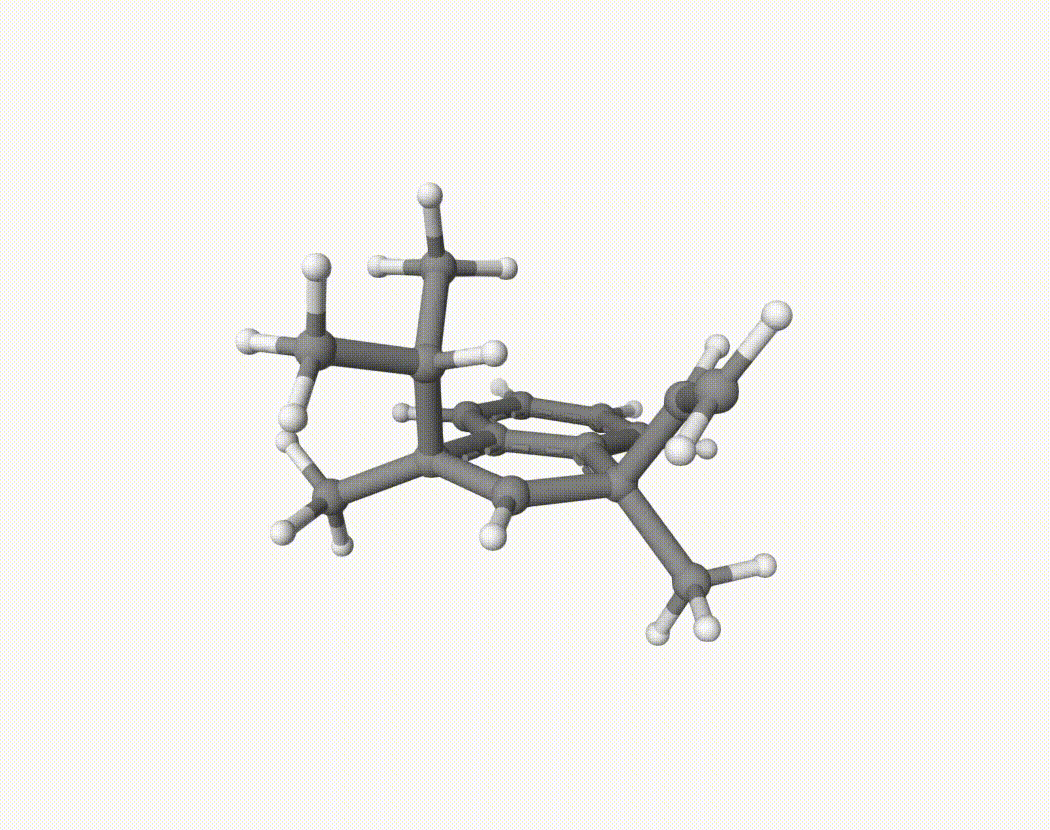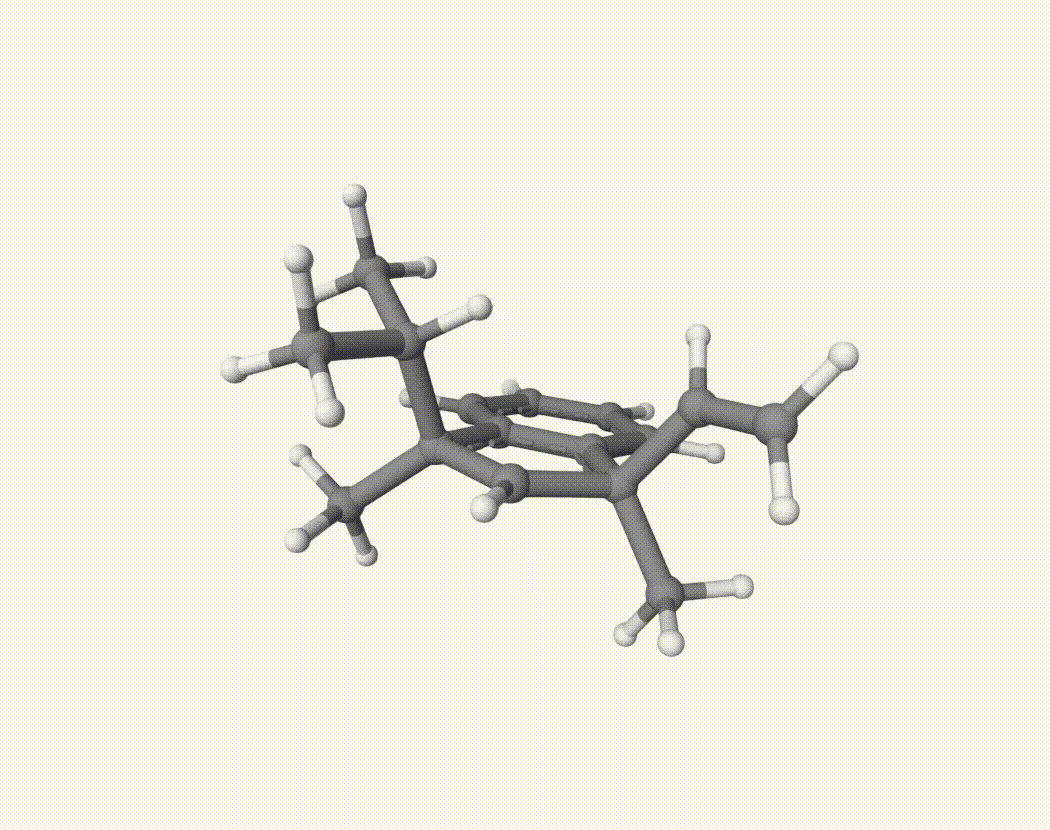
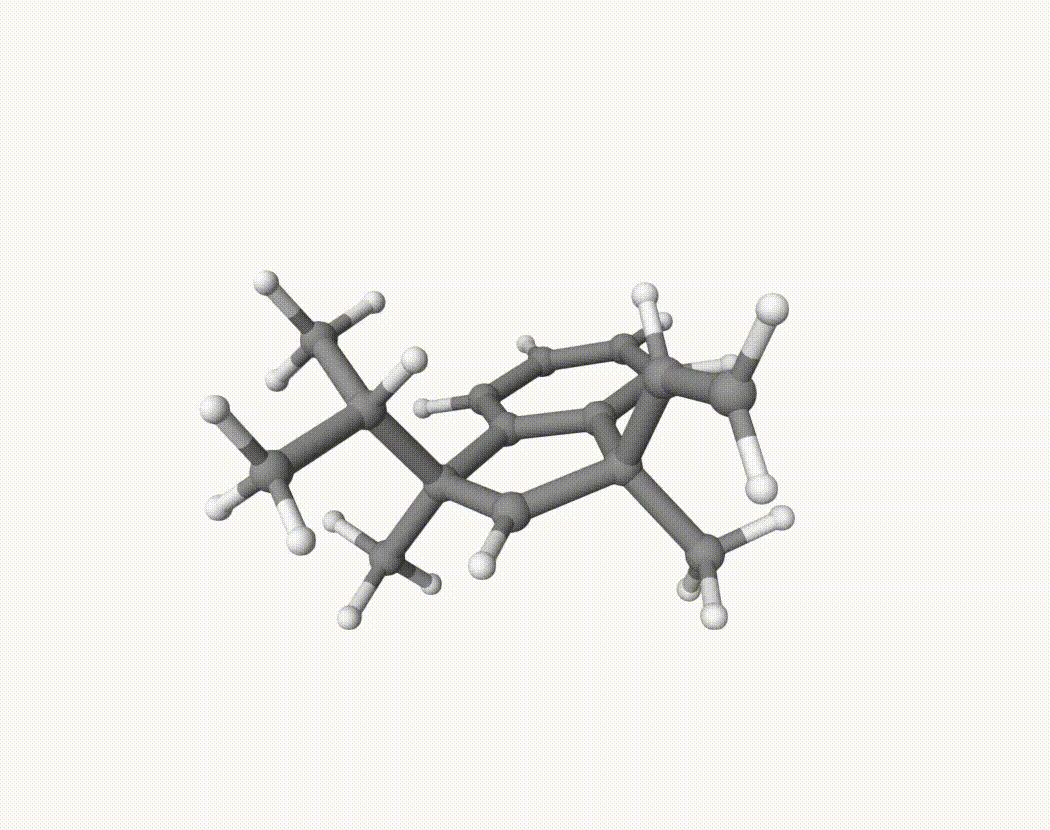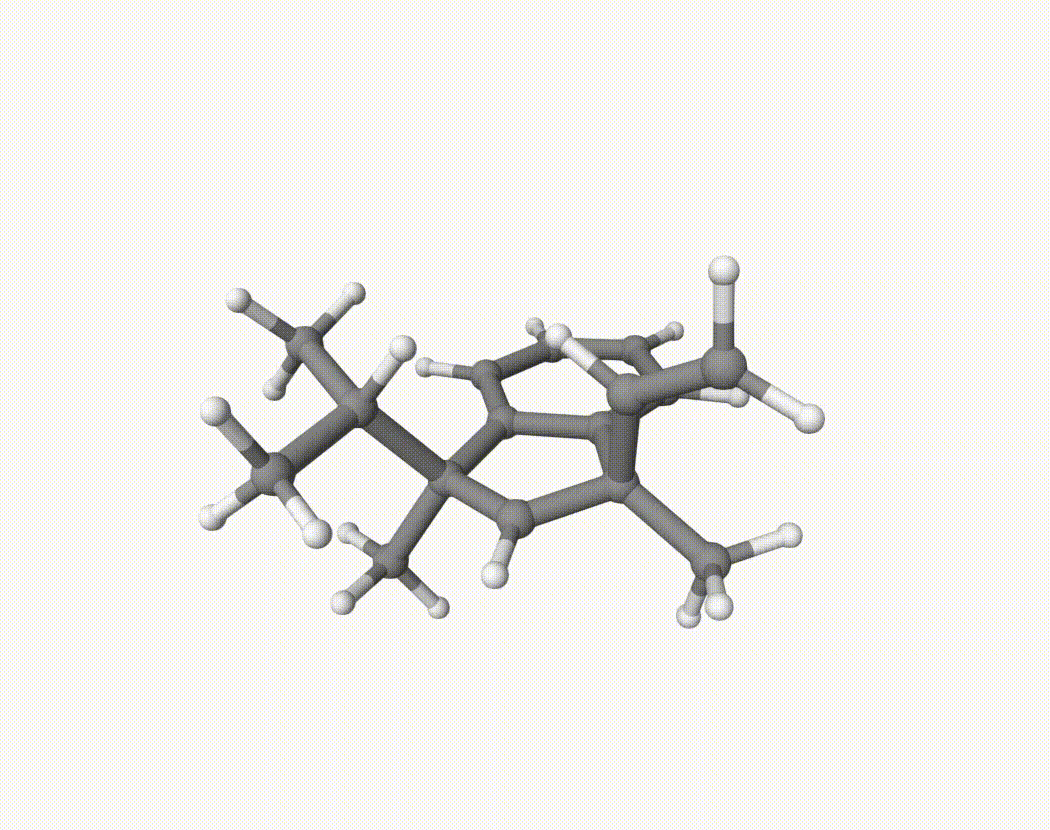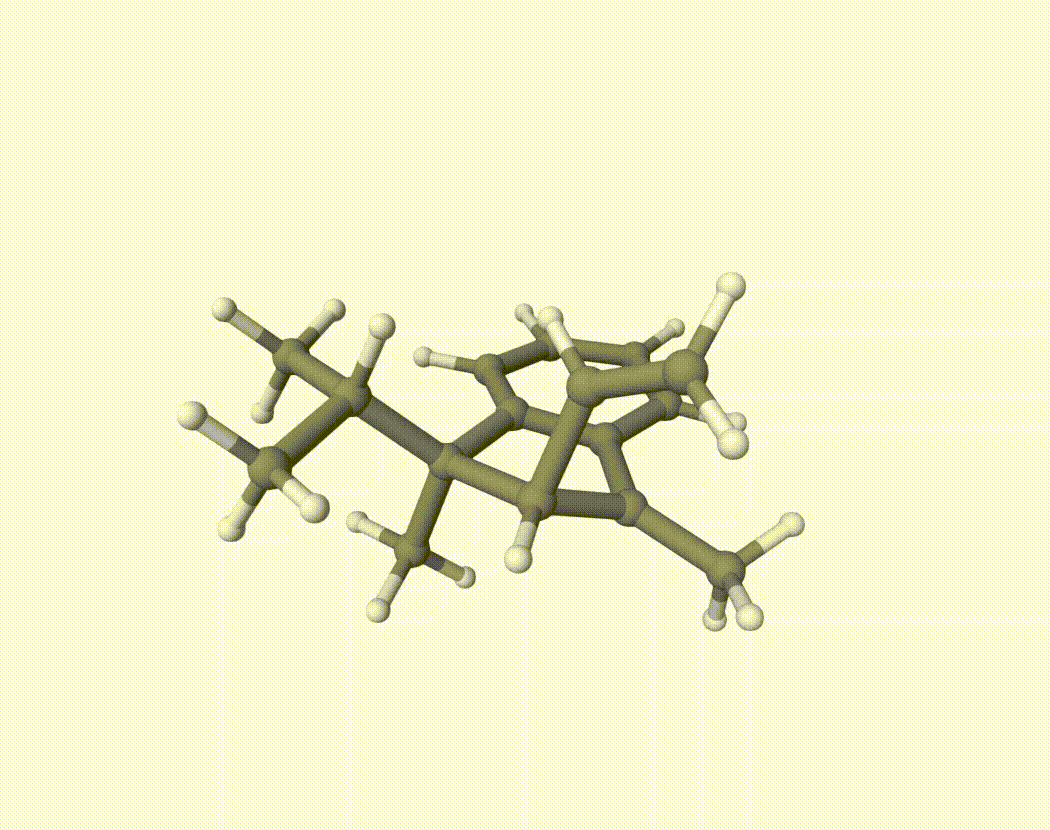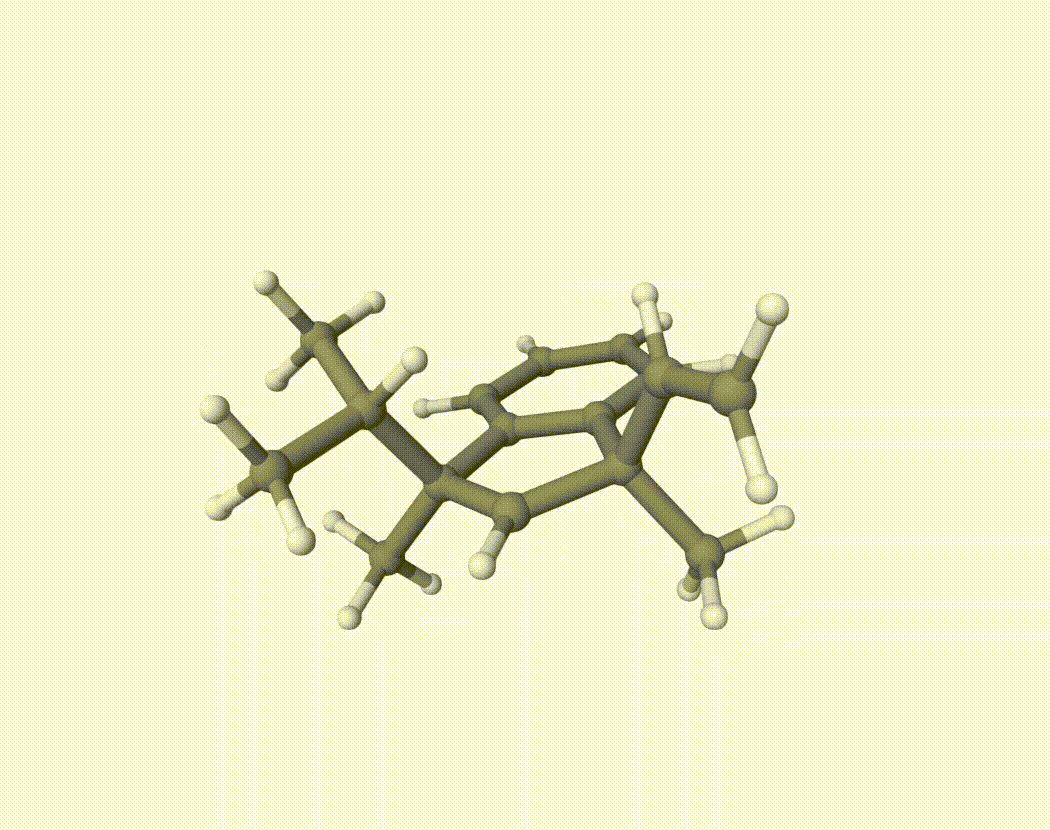
For this test, we can calculate the migration ratio of the isopropyl group to the vinyl group. For 20 trajectories, the result is:
Isopropyl group migration |
7 |
|---|---|
Vinyl group migration |
7 |
Non-reactive |
2 |
Other reactions |
4 |
Of course, if the simulation time is longer, the non-reactive trajectory may become reactive. Anyway, the NVT MD result suggests that the migration ratio is about 1:1.
Why do we do dynamics? Due to the nonstatistical effect of this reaction, we cannot predict the migration ratio by a static calculation. The dynamics result is more reliable. Please refer to the paper J. Am. Chem. Soc. 2021, 143, 1088 for more details.