Tip
All input files can be downloaded: Files.
Energy Decomposition Analysis
This tutorial will lead you step by step to do energy decomposition analysis (EDA) using Qbics. Currently, EDA can only be used with DFT.
Hint
If you use Qbics to do EDA in you paper, please cite the following references:
Example: EDA for (H2SO4)(NH3)
Here, we will do an EDA for the molecular dimer (H2SO4)(NH3). The input is given below:
1basis
2 def2-svp
3end
4
5scf
6 charge 0
7 spin2p1 1
8 type U # This is needed for EDA.
9end
10
11grimmedisp
12 type bj # Options: none, zero, bj.
13end
14
15eda
16 type tso
17 frag 0 1 1-7
18 frag 0 1 8-11
19end
20
21mol
22 S -0.62161000 -0.11181000 0.09167200
23 O 0.03138700 -0.14316200 1.35576500
24 O -1.82120800 -0.82692400 -0.15912300
25 O 0.38678100 -0.46814500 -1.02324300
26 O -0.91128600 1.43385100 -0.17168300
27 H 1.34667100 -0.25497400 -0.71202200
28 H -1.58729700 1.50230600 -0.85583700
29 N 2.78190500 0.03915200 -0.05091700
30 H 3.20716100 0.92239700 -0.30232700
31 H 3.46881800 -0.69082400 -0.19060100
32 H 2.55167500 0.07103700 0.93671700
33end
34
35task
36 eda b3lyp
37end
Line 8: For EDA, you must explicitly set the
typetoUinscf...end.Line 16: Do TSO-type EDA. You can also use
gksto do generalized Kohn-Sham (GKS) type EDA.Line 17-18: The fragments in EDA. Each line defines the charge, spin multiplicity, and atom indices. See below:
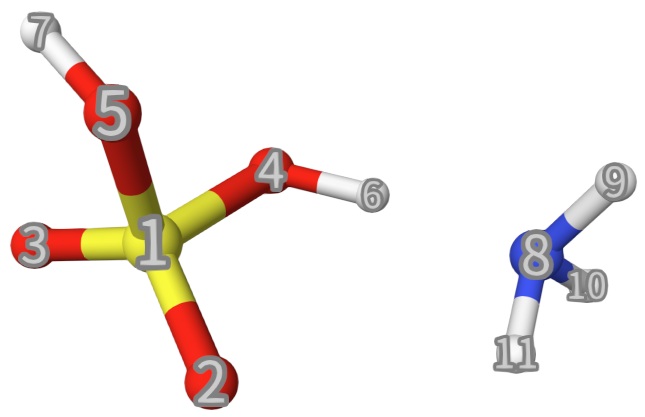
After running this calculation, we have the following output:
1WITHOUT BSSE correction:
2Electrostatic interaction energy: -30.95 kcal/mol
3Exchange-correlation interaction energy: 27.55 kcal/mol
4Polarization interaction energy: -5.44 kcal/mol
5Charge transfer interaction energy: -15.25 kcal/mol
6Grimme's dispersion interaction: -2.22 kcal/mol
7----------------------------------------------------------------
8Total interaction energy: -26.31 kcal/mol
9
10WITH BSSE correction:
11Electrostatic interaction energy: -30.95 kcal/mol
12Exchange-correlation interaction energy: 27.55 kcal/mol
13Polarization interaction energy: -5.44 kcal/mol
14Charge transfer interaction energy: -10.44 kcal/mol
15Grimme's dispersion interaction: -2.22 kcal/mol
16----------------------------------------------------------------
17Total interaction energy: -21.50 kcal/mol
In this output, the interaction energy of the following process
H2SO4 + NH3 = (H2SO4)(NH3)
is decomposed into several components. The interaction energy WITH and WITHOUT BSSE is -21.50 kcal/mol and -26.31 kcal/mol. The BSSE energy is included in Charge transfer interaction energy.
The Electrostatic interaction energy, Polarization interaction energy and Charge transfer interaction energy are all stablizing the dimer, the first contributing most. The Exchange-correlation interaction energy can also be recognized as Pauli exclusion energy, which destablizing the dimer.
For different kinds of molecular dimer, the weights of different components are quiet different, indicating the nature of chemical interactions.
Example: EDA for (Ca2+)(SO42-)(H2O)5
The basic MB-EDA formular is:
The terms higher than \(\Delta E^{(2)}\) is the many-body interaction term. Usually the most important one is the three-body effect \(\Delta E^{(3)}\), the effects of which can be decomposed into three ones:
\(\Delta E^{(3)} < 0\): Indicate a cooperative effect of the monomers in a cluster. The many-body interaction is stabilizing the cluster. This is often seen in hydrogen bonding clusters, like water clusters.
\(\Delta E^{(3)} > 0\): Indicate an anti-cooperative effect of the monomers in a cluster. The many-body interaction is destabilizing the cluster. This is often seen in a cluster of charged species, like ionic liquid clusters. Also see below.
\(\Delta E^{(3)} \approx 0\): Indicate a non-cooperative effect of the monomers in a cluster. There is little many-body interaction in the cluster. This is often seen in a cluster of molecules without charges or hydrogen bonds.
Each order can be decomposed into electrostatic, exchange, polarization, charge transfer, and dispersion terms:
Usually, electrostatic and exchange terms are highly additive, while polarization and charge transfer terms are non-additive. The dispersion term is always additive.
Here we will do an EDA for the microsolvated cluster (Ca2+)(SO42-)(H2O)5. Since there are more than 2 species, we will use many-body EDA (MB-EDA) to decompose the interaction energy components into contributions from different many-body level. Usually, contributions beyond three-body are very small, so we will truncate the computation at four-body.
The input is given below:
1basis
2 def2-svp
3end
4
5scf
6 charge 0
7 spin2p1 1
8 type U # This is needed for EDA.
9end
10
11grimmedisp
12 type bj
13end
14
15eda
16 type mb_tso
17 mb_level 4 # The many-body truncation level.
18 frag +2 1 1
19 frag -2 1 2-6
20 frag 0 1 7-9
21 frag 0 1 10-12
22 frag 0 1 13-15
23 frag 0 1 16-18
24 frag 0 1 19-21
25end
26
27mol
28 Ca 1.98604937 1.24582501 1.89905383
29 S 0.69103489 3.45226578 0.98237460
30 O 2.08708072 3.44779165 1.50308184
31 O 0.24826049 2.05102931 0.73622996
32 O -0.21032250 4.08940001 1.98323165
33 O 0.63912085 4.22084216 -0.29304506
34 O 3.02175028 0.83377391 -0.05175855
35 H 3.13194669 0.09601115 -0.65158745
36 H 2.90415260 1.59241194 -0.62348488
37 O 2.51307423 2.92493798 -1.72944079
38 H 1.80535522 3.44397361 -1.34738364
39 H 2.91500306 3.50585612 -2.37536585
40 O 0.60914213 -0.51387571 1.58896733
41 H 0.07645358 -1.26917200 1.83796991
42 H 0.01571971 0.04244367 1.08439778
43 O 0.73702072 1.93958625 3.70546127
44 H 0.73393089 2.03509947 4.65787899
45 H 0.20818330 2.67203308 3.38909995
46 O 3.87639195 1.93942131 2.91131853
47 H 4.78820918 1.95286631 3.20224881
48 H 3.69756418 2.83995160 2.64058309
49end
50
51task
52 eda b3lyp
53end
Line 16: To do MB-EDA, you should use
mb_tso.Line 17:
mb_level 4means you truncate the MB-EDA at four-body.Line 18-24: Set 7 chemical fragements. Note that some species like Ca2+ and SO42- are charged. You should give the correct charge.
The atomic indices are shown below:
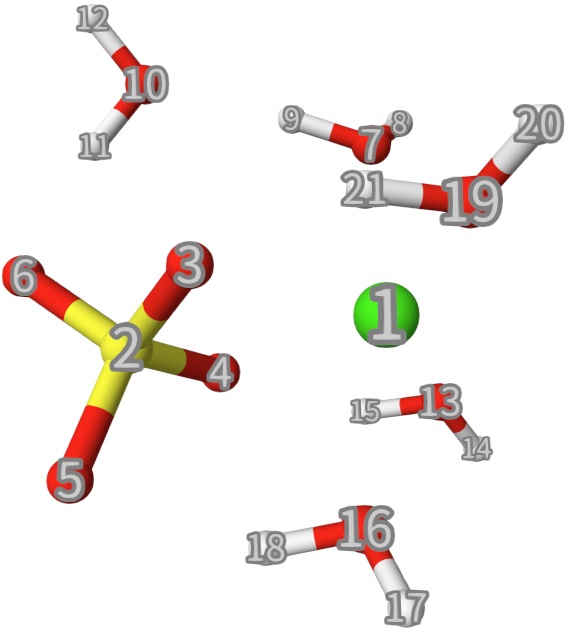
After running this calculation, we have the following output:
1Table 5. Summary (kcal/mol).
2---------------------------------------------------------------------------------------------------------------------------------
3 Interactions delE_el delE_xc delE_pl delE_ct delE_bsse delE_disp delE_tot
4---------------------------------------------------------------------------------------------------------------------------------
5 SUM of 2-body -6.73148230E+02 1.47696370E+02 -1.11395126E+02 -2.04547116E+02 1.03684957E+02 -2.06082308E+01 -7.58317375E+02
6 SUM of 3-body 1.42501290E-08 -2.67700179E+00 6.09855572E+01 1.30716087E+02 -4.86421459E+01 1.07484123E+01 1.51130908E+02
7 SUM of 4-body -2.82504918E-08 5.22296932E-01 -4.66678754E+00 -4.78862109E+01 1.47634652E+01 -1.52639538E+01 -5.25311901E+01
8 Remain 1.63009297E-08 -9.27025377E-02 2.21610464E-01 1.02616142E+01 -2.89029982E+00 7.47122033E+00 1.49714426E+01
9---------------------------------------------------------------------------------------------------------------------------------
10 SUM -6.73148230E+02 1.45448963E+02 -5.48547462E+01 -1.11455626E+02 6.69159769E+01 -1.76525519E+01 -6.44746214E+02
11---------------------------------------------------------------------------------------------------------------------------------
We can see that the total interaction energy is -644.75 kcal/mol, which is decomposed into 2-body, 3-body, 4-body, and remaining terms. The 2-body term is the most important one (-758.32 kcal/mol), while the 3-body term is also significant but anti-cooperative (destablizing the cluster) (+151.13 kcal/mol). The 4-body term is small (-52.53 kcal/mol, slightly cooperative). The remaining term (sum of 5-, 6-, and 7-body) is very small (+14.97 kcal/mol) compared to the low-order terms.
We can also see that the electrostatic and exchange energy are highly additive (beyond 3-body is almost zero), while the polarization and charge-transfer energy are non-additive:
Type |
2-body |
3-body |
4-body |
Total |
|---|---|---|---|---|
Polarization |
-111.40 kcal/mol |
+60.99 kcal/mol |
-4.67 kcal/mol |
-54.85 kcal/mol |
Charge transfer |
-204.55 kcal/mol |
+130.72 kcal/mol |
-47.89 kcal/mol |
-111.45 kcal/mol |
For different kinds of clusters, the 3-body effects (many-body interactions) can be quite different, see Phys. Chem. Chem. Phys. 2024, 26, 17549 for more information.