Tip
All input files can be downloaded: Files.
mecp
This keyword defines the arguments for searching minimum energy crossing point (MECP) and using diabatic MECP (dMECP) method to search transition states.
For standard MECP search, you can use the mecp keyword in task...end block. For using dMECP to search transition states, you can use the dmecp keyword in task...end block.
Hint
When using this keyword, in scf...end block, you must ALWAYS set type u.
Options
- num_steps
Value
An integer
Default
200
The maximum number of MECP search.
- energy_cov
Value
A real number
Default
1.E-5The energy difference convergence threshold. When
energy_cov,grad_cov, anddr_covare all met, the MECP will be set to be converged.
- grad_cov
Value
A real number
Default
1.E-3The gradient convergence threshold. When
energy_cov,grad_cov, anddr_covare all met, the MECP will be set to be converged.
- dr_cov
Value
A real number
Default
1.E-3The displacement convergence threshold. When
energy_cov,grad_cov, anddr_covare all met, the MECP will be set to be converged.
- orb1
This defines the electronic configuration of state 1. The format is:
orb num_electrons spin_multiplicity alpha_MO_indices : beta_MO_indicesThere can be arbitrary number of
orb1, but all orbitals must be included once and only once. They are the same asorbinscfguess. See scfguess.
- orb2
This defines the electronic configuration of state 2. The format is:
orb num_electrons spin_multiplicity alpha_MO_indices : beta_MO_indicesThere can be arbitrary number of
orb2, but all orbitals must be included once and only once. They are the same asorbinscfguess. See scfguess.
- frag1
This defines the fragmentation of state 1. The format is:
frag num_electrons spin_multiplicity atom_rangeThere can be arbitrary number of
frag1, but all atoms must be included once and only once. They are the same asfraginscfguess. See scfguess.
- frag2
This defines the fragmentation of state 1. The format is:
frag num_electrons spin_multiplicity atom_rangeThere can be arbitrary number of
frag2, but all atoms must be included once and only once. They are the same asfraginscfguess. See scfguess.Hint
When
frag1/2andorb1/2are defined simultaneously,orb1/2will be considered in priority.
Theoretical Background
MECP Between Ground and Excited States
The minimum energy crossing point (MECP) is a point where two potential energy surfaces (PES) of different electronic states cross each other. In the context of quantum chemistry, it is often used to describe the transition between a ground state and an excited state. The MECP can be found by optimizing the geometry of the system while constraining the energy difference between the two states to be zero.
Diabatic MECP (dMECP)
Diabatic MECP (dMECP) is a method used to find the transition state for a AB+C=A+BC reaction. It uses the MECP search algorithm to find the minimum energy crossing point between the reactant and product diabatic states.
Input Examples
Example: MECP between the Singlet and Triplet States of C6H5+
In this example, we will search the MECP between the singlet and triplet states of C6H5+. The input file is as follows:
1basis
2 def2-svp
3end
4
5mol
6 C 0.00000000 1.39661300 0.00000000
7 C 1.20950300 0.69830700 0.00000000
8 C 1.20950300 -0.69830700 0.00000000
9 C 0.00000000 -1.39661300 0.00000000
10 C -1.20950300 -0.69830700 0.00000000
11 C -1.20950300 0.69830700 0.00000000
12 H 2.15088200 1.24181200 0.00000000
13 H 2.15088200 -1.24181200 0.00000000
14 H 0.00000000 -2.48362500 0.00000000
15 H -2.15088200 -1.24181200 0.00000000
16 H -2.15088200 1.24181200 0.00000000
17end
18
19mecp
20 num_steps 200 # The number of search steps.
21 energy_cov 1.E-5 # The energy difference convergence threshold.
22 orb1 40 1 1-109 : 1-109
23 orb2 40 3 1-109 : 1-109
24end
25
26scf
27 type u
28 charge +1
29 spin2p1 1
30end
31
32task
33 mecp b3lyp
34end
In mecp...end block, we define the two electronic configurations of the singlet and triplet states using orb1 and orb2. The num_steps is set to 200, and the energy convergence threshold is set to 1.E-5.
The format of orb1 and orb2 is the same as that in scfguess. For example, orb1 40 1 1-109 : 1-109 means that the first state has 40 electrons, a spin multiplicity of 1, and the alpha and beta MOs are from indices 1 to 109. The same applies to orb2.
The singlet-triplet MECP is saved to mecp-1-mecp.xyz. The optimized process is saved in mecp-1-mecp-traj.xyz, which can be shown using Qbics-MolStar:
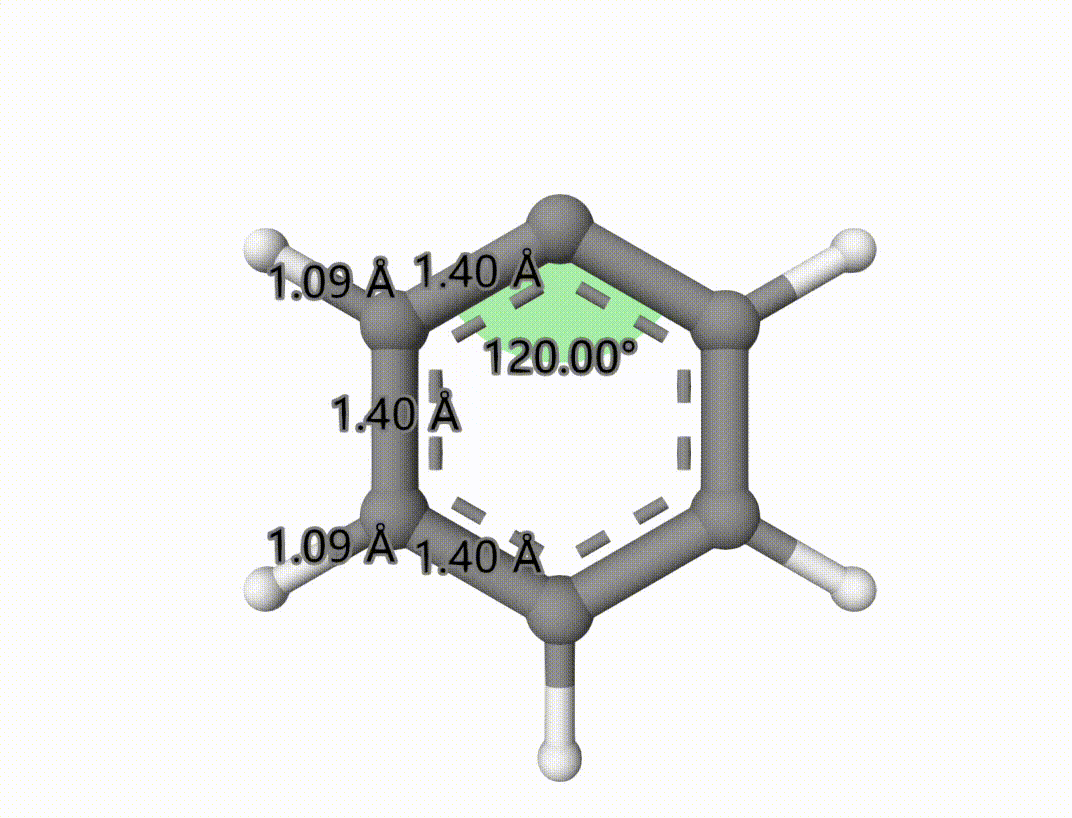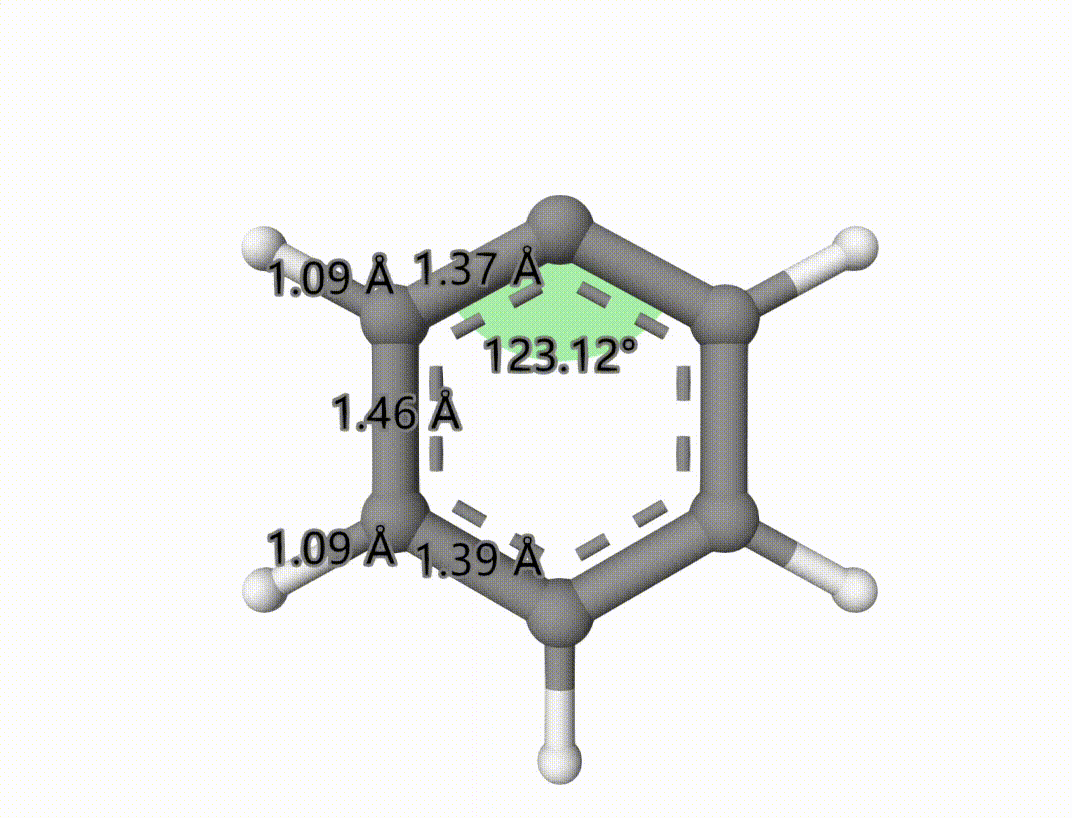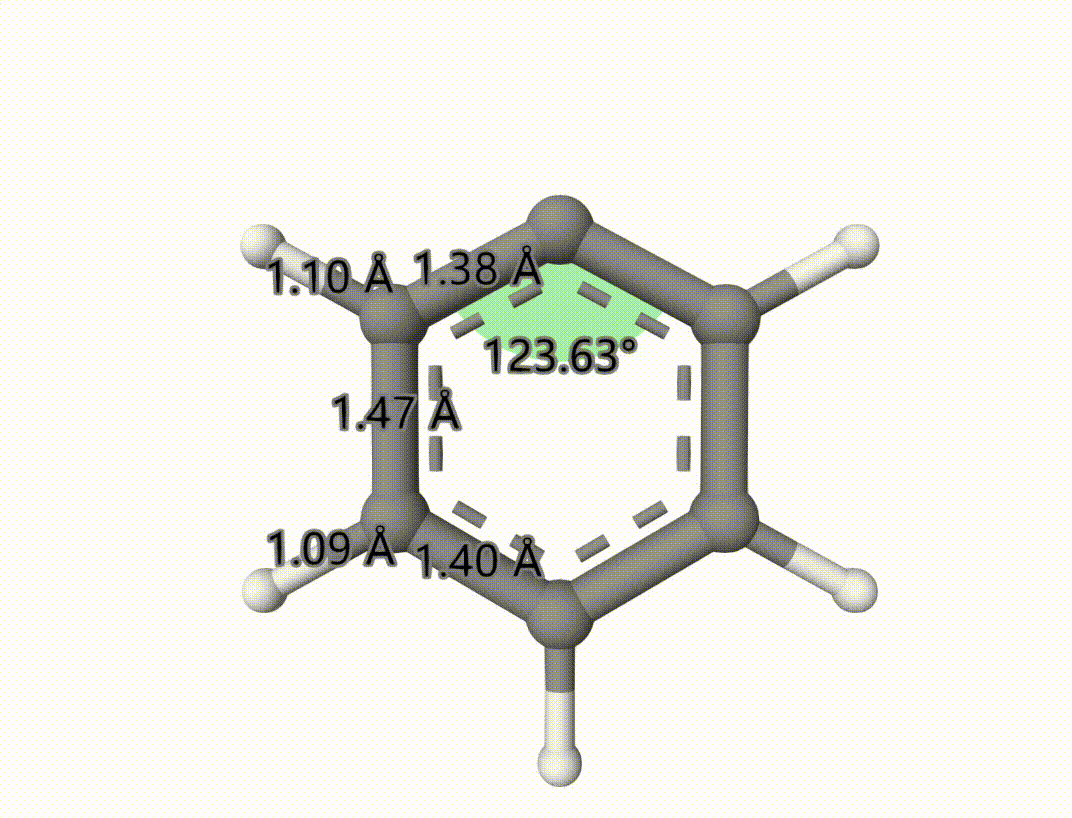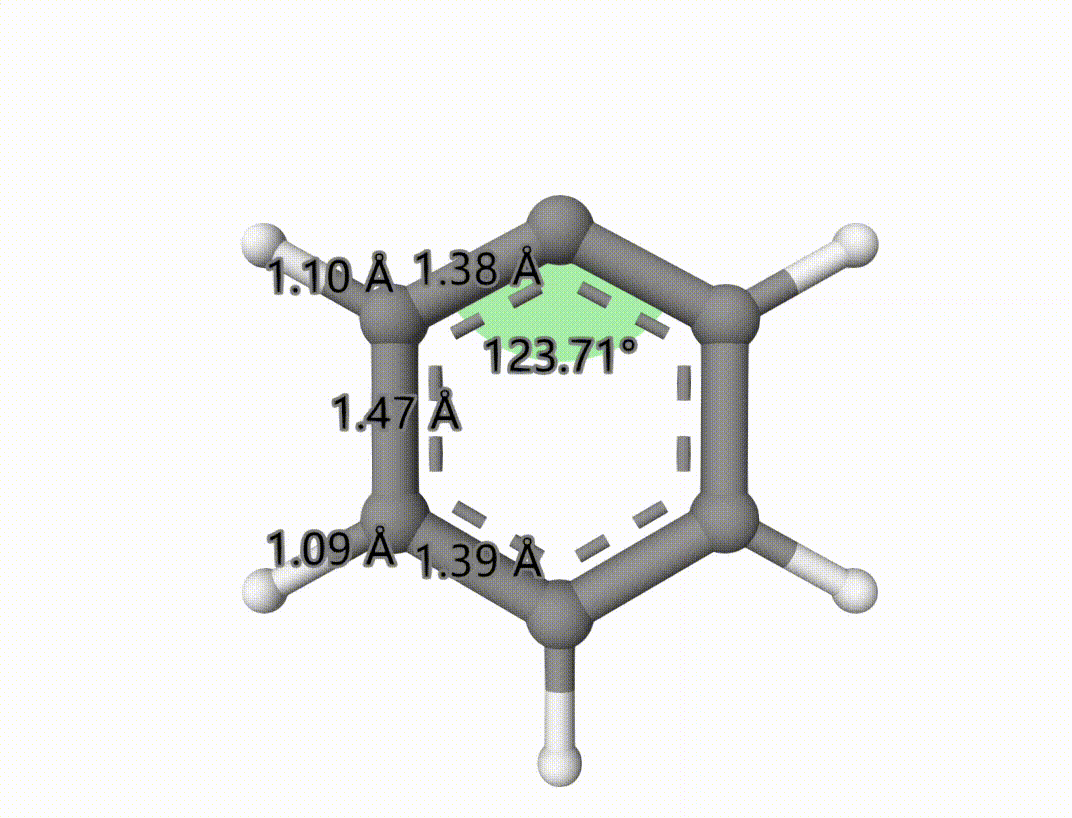
Example: Search Transition State of Iodine Atom Transfer Reaction
Hint
To use dMECP, you must use the dmecp keyword in task...end block instead of mecp. The obtained structures using dmecp and mecp are different. For transition states, we recommend to use ``dmecp`` instead of ``mecp`` to search the transition state.
In this section, we want to search the transition state of the following reaction:
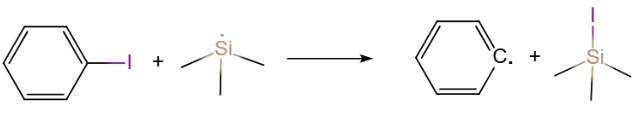
This is a AB+C=A+BC type reaction. The largest advantage of dMECP algorithm is that the transition state is searched using energy coordinate instead of geometry coordinate, so it is not too sensitive to the initial guess.
The input file is as follows. Note that for iodine, we have used pseudopotential with pseudopotential keyword, def2-ecp.
1basis
2 def2-svp
3end
4
5pseudopotential
6 def2-ecp
7end
8
9scf
10 type u
11 charge 0
12 spin2p1 2
13end
14
15mol
16 C -0.29500 -0.00002 -0.00017
17 C 1.10016 -0.00002 -0.00017
18 C 1.79770 1.20773 -0.00017
19 C 1.10004 2.41624 -0.00137
20 C -0.29478 2.41616 -0.00185
21 C -0.99238 1.20795 -0.00085
22 H -0.84476 -0.95234 0.00028
23 H 1.64967 -0.95253 0.00114
24 I 3.87770 1.20788 0.00103
25 H 1.65024 3.36838 -0.00143
26 H -0.84490 3.36844 -0.00280
27 H -2.09199 1.20814 -0.00103
28 Si 6.95770 1.20810 0.00280
29 C 7.59353 0.30695 1.56221
30 C 7.59533 0.30859 -1.55681
31 C 7.59423 3.00892 0.00412
32 H 7.23873 -0.70024 -1.55754
33 H 7.23913 0.81342 -2.43040
34 H 8.66533 0.30866 -1.55619
35 H 8.66353 0.30703 1.56282
36 H 7.23632 0.81087 2.43592
37 H 7.23694 -0.70188 1.56147
38 H 8.66423 3.00899 0.00563
39 H 7.23876 3.51349 -0.86992
40 H 7.23629 3.51309 0.87738
41end
42
43mecp
44 num_steps 200
45 energy_cov 1.E-5
46 frag1 0 1 1-12
47 frag1 0 2 13-25
48 frag2 0 2 1-8 10-12
49 frag2 0 1 9 13-25
50end
51
52task
53 dmecp b3lyp # Using dMECP method.
54end
The structure given in mol...end is shown below:
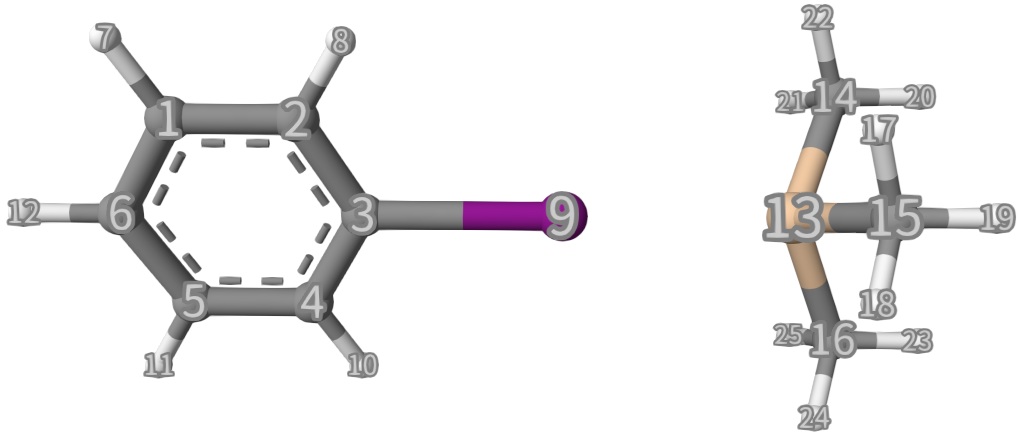
We can see that the initial structure is a complex that two reactants are simply putting together. Now, according to the reactant and product species, we can define frag1 and frag2 in mecp...end option block. The definitions are shown below, which is self-explanatory and in line with chemical intuition:
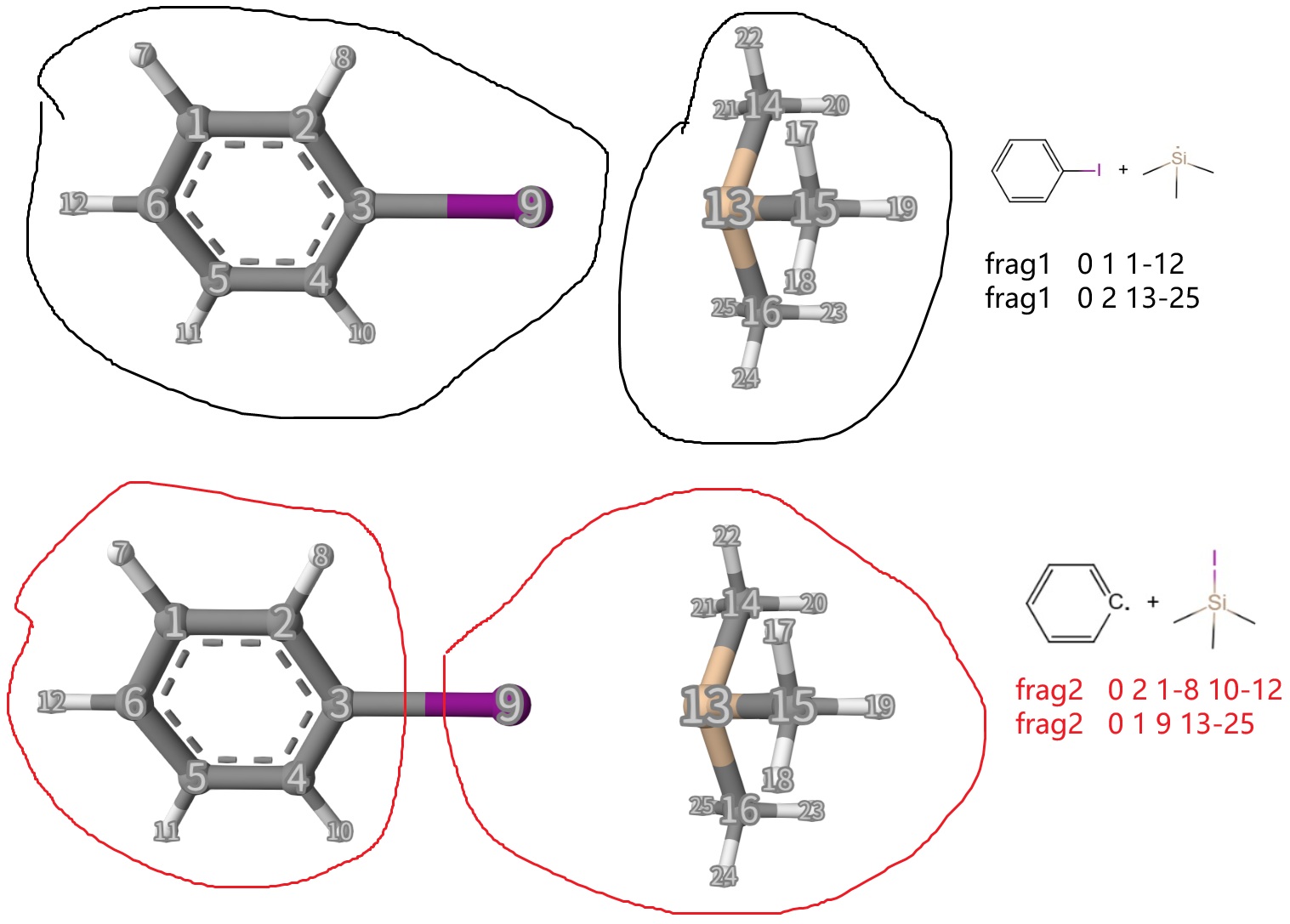
In dMECP method, only a single geometry is needed. Of course, the closer it approaches the transition state, the better.
You can also use mecp keyword instead of dmecp to to search the real MECP between the two diabatic states, the input files being as follows:
1basis
2 def2-svp
3end
4
5pseudopotential
6 def2-ecp
7end
8
9scf
10 type u
11 charge 0
12 spin2p1 2
13end
14
15mol
16 C -0.29500 -0.00002 -0.00017
17 C 1.10016 -0.00002 -0.00017
18 C 1.79770 1.20773 -0.00017
19 C 1.10004 2.41624 -0.00137
20 C -0.29478 2.41616 -0.00185
21 C -0.99238 1.20795 -0.00085
22 H -0.84476 -0.95234 0.00028
23 H 1.64967 -0.95253 0.00114
24 I 3.87770 1.20788 0.00103
25 H 1.65024 3.36838 -0.00143
26 H -0.84490 3.36844 -0.00280
27 H -2.09199 1.20814 -0.00103
28 Si 6.95770 1.20810 0.00280
29 C 7.59353 0.30695 1.56221
30 C 7.59533 0.30859 -1.55681
31 C 7.59423 3.00892 0.00412
32 H 7.23873 -0.70024 -1.55754
33 H 7.23913 0.81342 -2.43040
34 H 8.66533 0.30866 -1.55619
35 H 8.66353 0.30703 1.56282
36 H 7.23632 0.81087 2.43592
37 H 7.23694 -0.70188 1.56147
38 H 8.66423 3.00899 0.00563
39 H 7.23876 3.51349 -0.86992
40 H 7.23629 3.51309 0.87738
41end
42
43mecp
44 num_steps 200
45 energy_cov 1.E-5
46 frag1 0 1 1-12
47 frag1 0 2 13-25
48 frag2 0 2 1-8 10-12
49 frag2 0 1 9 13-25
50end
51
52task
53 mecp b3lyp # Search MECP between the two diabatic states.
54end
The initial and optimized structures mecp-2*.inp, mecp-2a-mecp.xyz and mecp-2b-mecp.xyz are shown below:
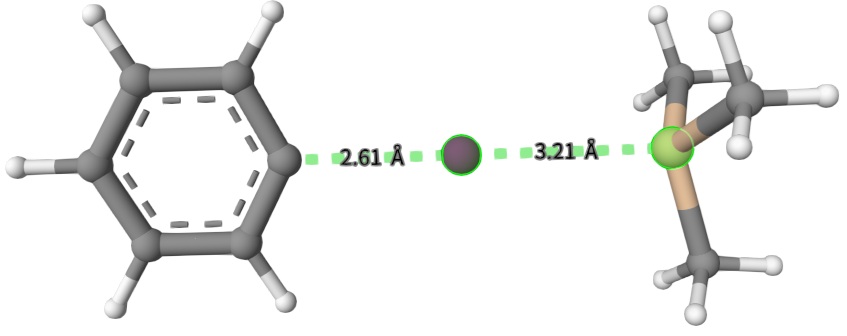
Interestingly, both mecp-2a-mecp.xyz and mecp-2b-mecp.xyz have a single imaginary frequency, indicating that they are both close to transition states. However, the transition state obtained by dmecp is more stable than that obtained by mecp. For mecp-2b-mecp.xyz, we find that the energy difference between the reactant and product diabatic states is about 0.000008 Hartree, which is the definition of MECP. For transition states, we recommend to use ``dmecp`` instead of ``mecp`` to search the transition state.